Research efforts led by Joshua Denny, M.D., director of Vanderbilt’s Center for Precision Medicine, are leveraging the rich data in EHRs for large-cohort analyses of genome-phenome associations. Results are both confirming and revealing new relationships to disease development and drug response.
Discoveries: Before we discuss the new study of HLA variations published in Science Translational Medicine, would you explain the PheWAS methodology that is informing your work at the Center for Precision Medicine?
Denny: PheWAS stands for phenome-wide association study. It’s based on the premise that as we see patients in the clinic, we record many observations that become part of their medical record. As we collect really dense EHR data, we have the opportunity not just to study one given disease or phenotype, but really to look across many or all the phenotypes one may accrue through the medical record.
When I define the phenome, I’m thinking about the clinical diseases, signs and symptoms that can be studied in an individual patient. We look at it across a population. We are using PheWAS to help explain genetic findings and really understand the genetic underpinnings of disease.
Discoveries: Would you say more about the sources of data empowering this type of study?
Denny: Vanderbilt began an early investment in EHRs for clinical care, just like everyone else. But we built it thinking about chances to build a repository for this information, which could help us get smarter in healthcare. Part of that is researching and learning more about the population with every patient you see.
We have about 2.5 million patients within our EHR at Vanderbilt. We began a collection to link these patients to their DNA with their consent, such that when a patient gives a normal blood sample for a normal, routine clinical test. Some of that leftover blood could be banked for DNA research. Now we have a population of about 250,000 people for which we have DNA. We can study the whole range of clinical phenotypes that they have against their DNA and learn what genetics predict disease, and what genetics may predict why a drug works or doesn’t work.
“We have a very rich data set that is almost a free byproduct of clinical care.”
Discoveries: How does this research work differently from a traditional study?
Denny: What’s great about this is that we have a very rich data set that is almost a free byproduct of clinical care. If you look at observational trials or research studies that are using clinical trial data, most studies look at one disease or a collection of related diseases. We’re able to look at a single population across potentially thousands of phenotypes. The PheWAS we typically do is around 1,600 phenotypes.
Discoveries: What about the most recent study published in Science Translational Medicine?
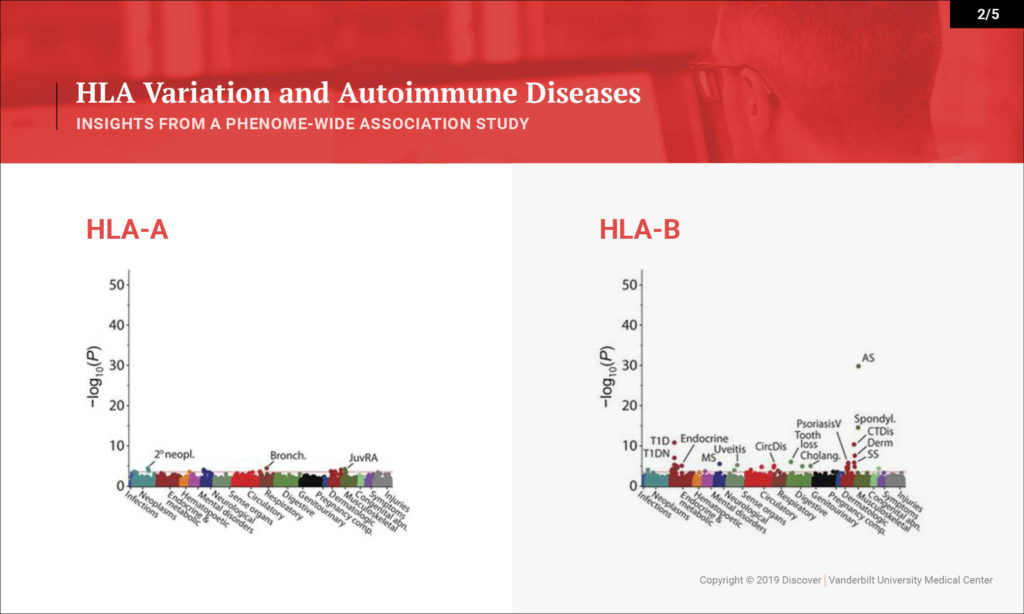
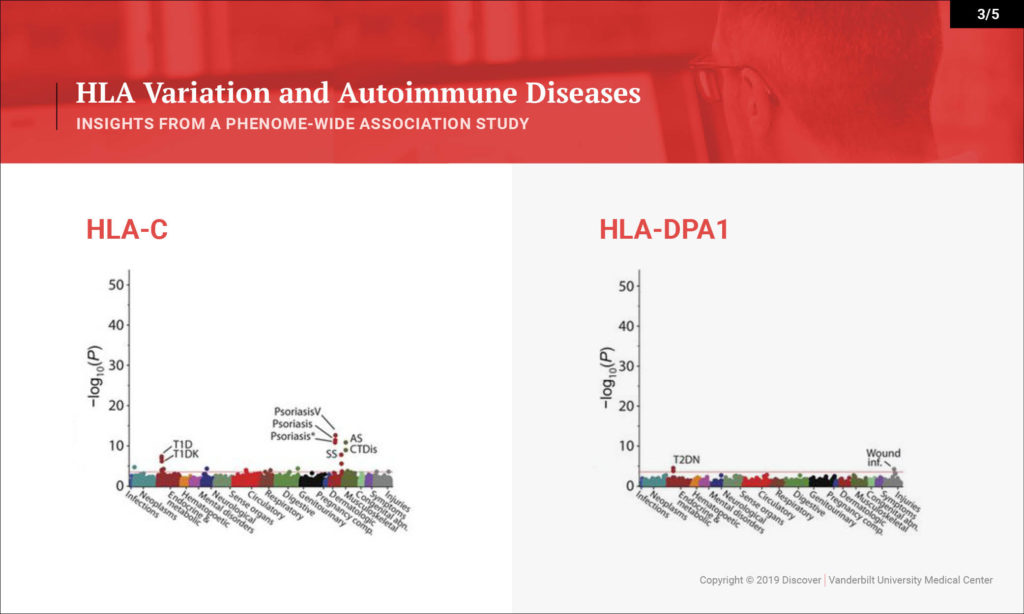
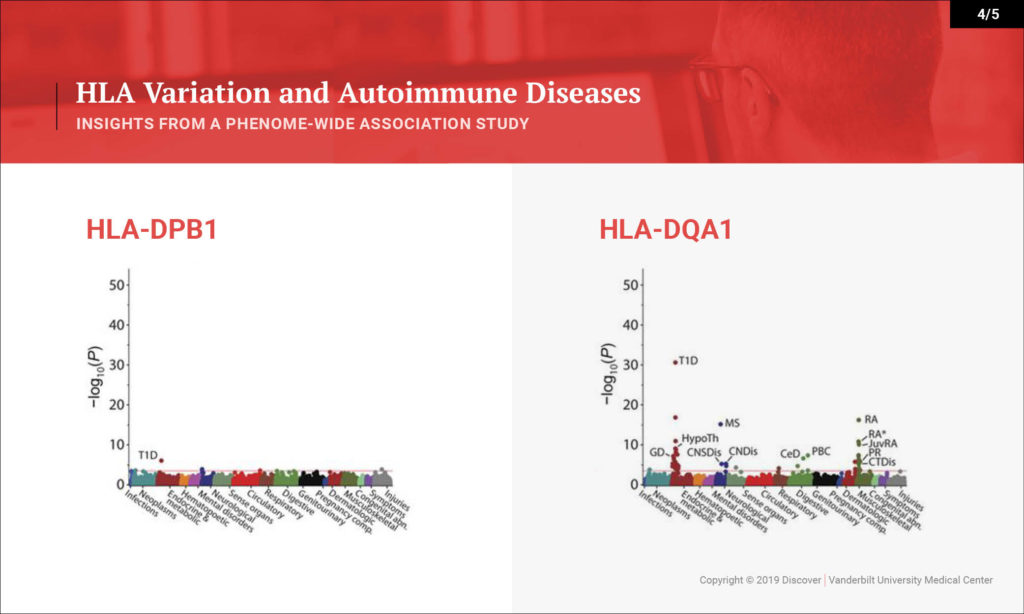
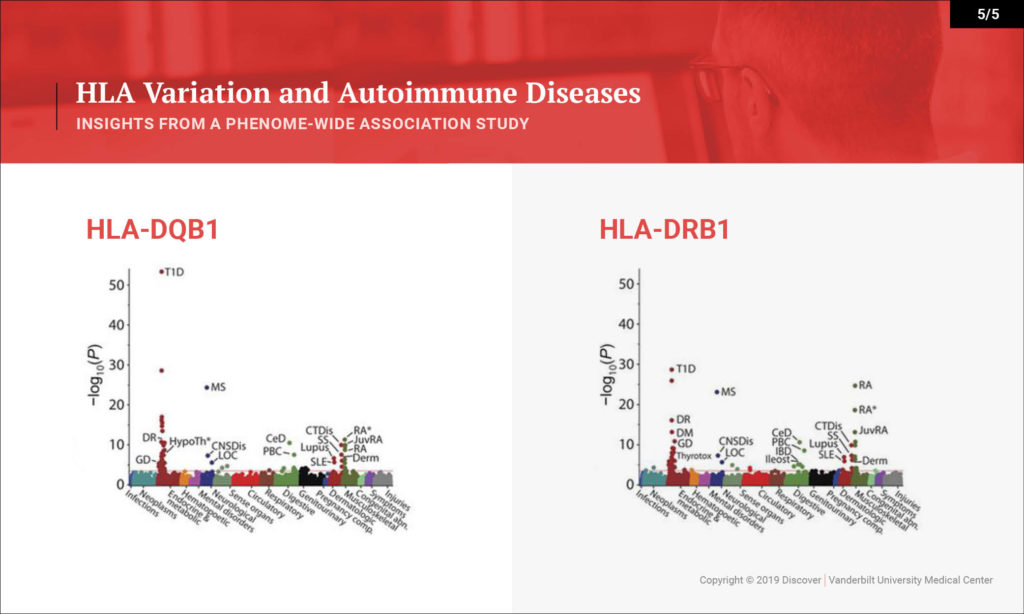
Denny: This study looked at the hypothesis that the HLA region, which is one of the most variable regions in the genome, is associated with many types of diseases. We know that it underlies much of autoimmune diseases and many rheumatologic diseases. That hasn’t been studied in one population across all the autoimmune diseases you can study and across the diversity of diseases that people face.
What we sought to do is investigate a large population — 37,000 people with DNA linked to their EHRs — and to study the impact of variation within the HLA region on their diseases. We were able to replicate many of the known findings from the last several decades very quickly in this population. We replicated over 100 findings, most of which are known, but we were also able to explore that existing knowledge in more depth. We discovered a few new findings as well.
Discoveries: When you look for associations between the HLA variant and specific diseases, what are you really looking for? What is the methodology?
Denny: We start with DNA and genotype them. We’re pretty high-depth, and from there we figure out what we call the HLA type. We can also figure out the amino acids behind the HLA type, specifically for the immunoglobulins. We take that data and are able to compare it against their diseases. We look across all the diseases that are part of the chart, and we identify cases and controls for each of these 1,600 diseases. Then we study them, doing association tests to look for significant association.
Discoveries: What are some exciting findings in that recent research?
Denny: On one hand, we’re able to rapidly provide evidence for a lot of what’s been known already. One interesting byproduct is the ability to look at disease comorbidity. Any rheumatologist or clinician will see patients that have type 1 diabetes and rheumatoid arthritis. We know these clinically occur together. Is that because they share genetics, or is that because one disease may actually put someone at risk for the other? Or maybe the treatment for one disease puts one at risk for the other. We can figure that out with genetics. This study tells us that for the most part, having the genetics puts us at risk for both autoimmune diseases.
Some of our new findings point toward HLA types indicating potentially more severe disease – more severe types of type 1 diabetes with their complications, as opposed to less severe type 1 diabetes. That could be through earlier onset. That could be through more aggressive disease itself, or more difficulty in controlling glucose. What we can show from this study is that certain subtypes of HLA seem to be associated with greater risk for certain severity of type 1 diabetes. For instance, there was also an association with cervical dysplasia, which is a predecessor for cervical cancer, which is novel.
Discoveries: As you’re identifying these associations between the genetic underpinnings and different diseases, what may be the next steps for research and developing new therapies?
Denny: A study like this can point to better understandings of risk for a given individual to develop other diseases, and maybe what their prognosis is. This particular study doesn’t give much insight yet for a new therapy. It could certainly lead toward new therapies.
Relatives of this study and future research using EHRs could further understanding of why a treatment works or doesn’t.
Discoveries: As director of the Center for Precision Medicine and a collaborator with peer institutions around the country, what areas of research have you feeling most excited?
Denny: I’m really excited about this paradigm of using passively-collected clinical data on massive scales with all the available genetic data and other biomarkers. We’ve seen an explosion of this kind of research in the last couple years. National efforts are really accelerating.
The All of Us Research Program in the U.S. is one of the more ambitious programs around the world. It seeks to enroll a million people who will be engaged partners in this research process and share biospecimens. Participants share their EHR data, answer health surveys and are available to participate in future clinical trials and other research studies. This could have a really profound impact. With our current study including only 37,000 people, imagine when we have a million people and can look across that population.
Discoveries: What partner organizations are helping to expand this type of research?
Denny: There are a number of sites across the country participating in the All of Us research program. Another network is the Electronic Medical Records and Genomics network (eMERGE), which really helped pioneer the use of EHRs for genomic research. We’re part of other networks doing this research as well, such as looking at the genetics of drug response or how to discover using EHR data, genetics and biomarkers.
Discoveries: At the level of an individual patient who might have rheumatoid arthritis, how will this type of research transform their care?
Denny: This paradigm of creating therapies against new and biologic targets is really a powerful one. Genetics is really helping foster that paradigm. We now have drugs coming on the market that are completely because of genetic discoveries. That is only going to accelerate and have a profound impact on autoimmune disease, just as it has in cardiovascular disease and some of the novel therapeutics we have in rheumatologic disease.
Another thing that will come is knowing better which medicine to start with, the right regimen and which side effects a patient might get. We’ll be able to customize therapy better. We may understand who’s at risk of severe comorbid disease like myocardial infarction, which is more common in a lot of autoimmune disease, and why that patient is more at risk. How could we alter that therapy? How could we alter that risk?
Discoveries: In addition to EHR and genetics, what other types of interesting research do you see here at Vanderbilt?
Denny: We understand that genetics drives part of disease, but what people do, their habits and their exposures are big parts, too. For instance, we’re learning ways of mining EHR data for textual cues to determine who has had an abuse episode in the past, who has exposures and may have been homeless or is at risk for homelessness. We’re looking at predictors for who is at risk for suicide. We’re understanding exposures such as smoking and alcohol. Those are all important risks of disease.
Another element is environmental exposures. We carry smartphones in our pockets that know where we are all the time. Advertising and internet companies are using that to pinpoint ads and know traffic patterns. We could use that to know more about exposures and correlate that with data collected by the EPA, or NOAA or other government organizations that give us information about access to high-quality foods, pollution status or how much green space is near you to [accommodate] exercise. We’ll be able to study those kinds of things in more granular ways, combined with genetics and health data, to understand better a person’s trajectory of health over time.