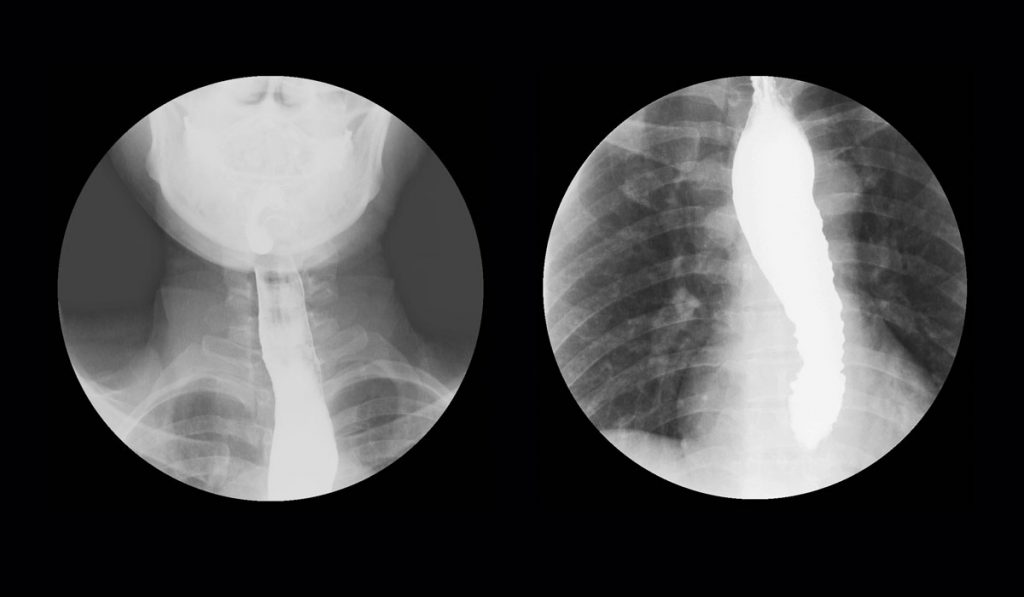
There has been significant evolution of the management of achalasia with the introduction of high-resolution manometry (HRM). However, limited knowledge about the etiology of the disease continues to fuel diagnostic and therapeutic challenges. Ongoing research by investigators at Vanderbilt University Medical Center is seeking to reveal biomarkers that will help clarify the disease mechanisms of achalasia.
“While achalasia is relatively rare, there several subtypes of the disease and each subtype behaves differently in terms of treatment and response,” said Rishi Naik, M.D., assistant professor of medicine in the Division of Gastroenterology, Hepatology and Nutrition at Vanderbilt. “Our hypothesis is that changes in inflammatory markers, genetic associations, and histologic and molecular signatures of proteins, lipids, and metabolites will define disease mechanisms based on clinical phenotypes.”
Growing Prevalence
Achalasia is a primary motor disorder characterized by impaired relaxation of the lower esophageal sphincter and by absence of peristalsis along the esophageal body. As a result, patients typically present with dysphagia, regurgitation and occasionally with chest pain, pulmonary complication and malnutrition.
Achalasia affects one to two people per 100,000 globally, but these numbers have grown in recent years based on more advanced diagnostics.
Searching for Biomarkers of Achalasia
Using Vanderbilt patient data, Naik’s team compared esophageal myotomy specimens collected during surgery against controls to: 1) define and compare inflammatory markers and histopathologic abnormalities within the esophageal myenteric plexus, and 2) identify differences in biomolecular composition of nerve tissue in the myenteric plexus. “Our goal was to combine proteomics, pathology and physiological data to identify the genes or proteins expressed in achalasia that are different from a normal person’s esophagus,” Naik said.
State-of-the-art imaging mass spectrometry identified changes in protein, lipid, and metabolite profiles in nerve and surrounding tissue from esophageal specimens. Spatially resolved tissue extraction combined with high sensitivity liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis was then performed. Shotgun proteomics was performed on the imaging mass spectrometry data.
“We’re hopeful this will uncover a personalized approach to treating patients.”
“For some time, we have thought there may be an autoimmune or viral component to achalasia,” Naik said. “We wanted to dig deeper and look at achalasia at the cellular level to understand why these patients are getting the disease.”
Finding Distinctions
Direct tissue examination showed important biochemical differences between achalasia and healthy esophageal specimens both via imaging mass spectrometry (ceramide phosphate, myelin basic protein) and immunohistochemistry (aganglionosis, venulitis). Histological analysis identified significant changes within achalasia subtypes, including a lower number of myenteric ganglion cells. Protein and lipids were specifically localized to nerve tissue and altered in achalasia tissue compared to controls.
The team will next perform genome and phenome studies in the esophageal samples and measure the abundance of varicella zoster virus DNA, transcripts and protein. The studies could help distinguish between specific achalasia subtypes.
“Identifying the biochemical profiles of these subtypes will help identify the etiology of achalasia,” Naik said. “All current measures of treatment are based on disrupting the lower esophageal sphincter; we’re hopeful that understanding why it gets tight will help us uncover personalized therapeutic approaches to treating patients from their genetic fingerprint.”